在某个普普通通的一天,我使用Seurat处理数据,但是发现这个数据却有点不那么普通
library(Seurat)
expr_matrix <- Read10X("/data5/ofo_data/yc/filtered_feature_bc_matrix_yc4/")
# 如果你的数据是一个矩阵或数据帧
rownames(expr_matrix) <- gsub("_", "-", rownames(expr_matrix))
seu <- CreateSeuratObject(counts = expr_matrix, project = "example")
seu <- subset(seu, subset = nFeature_RNA > 500 & nFeature_RNA <12000 & nCount_RNA >1000 & nCount_RNA <70000)
seu <- NormalizeData(seu)
seu <- FindVariableFeatures(seu, selection.method = "dispersion", nfeatures = 1000)
all.genes <- rownames(seu)
seu <- ScaleData(seu, features = all.genes)
seu <- RunPCA(seu, features = VariableFeatures(object = seu), npcs=50)
令人震惊的是,他提示了一个我从未见过的警告,我并没有在意,继续处理数据
Warning in irlba(A = t(x = object), nv = npcs, ...) :
You're computing too large a percentage of total singular values, use a standard svd instead.
Warning: Number of dimensions changing from 50 to 10
Warning messages:
1: Requested number is larger than the number of available items (11). Setting to 11.
2: Requested number is larger than the number of available items (11). Setting to 11.
3: Requested number is larger than the number of available items (11). Setting to 11.
4: Requested number is larger than the number of available items (11). Setting to 11.
5: Requested number is larger than the number of available items (11). Setting to 11.
然后我在构建SNN这一步,我就出错了,提示,给的维度太多了了。确实根据之前的提示信息,Seurat只算了10个PC的样子。
Error in FindNeighbors.Seurat(pbmc, dims = 1:50) :
More dimensions specified in dims than have been computed
于是我修改参数,把后续的维度改成了10
seu <- FindNeighbors(seu, dims = 1:10)
seu <- RunUMAP(seu, dims = 1:10)
seu <- FindClusters(seu, resolution = 0.5)
DimPlot(object = seu, reduction = "umap",label = TRUE,label.size = 7,repel = TRUE,pt.size = 0.3) +
theme(panel.grid = element_blank())
我得到了一个非常抽象的结果,我都不知道该如何解读
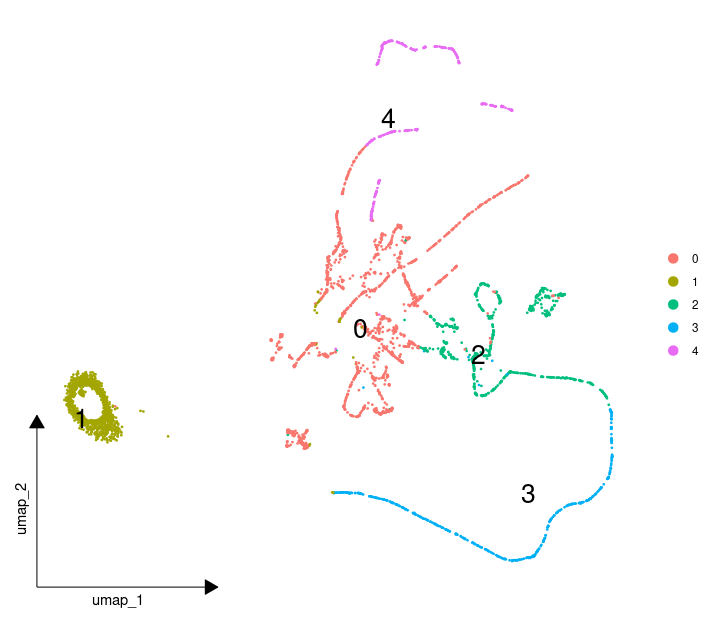
我推测一定是PCA这一步出了问题,我于是打算用警告里面提到可以用svd的方法。但是RunPCA 并没有选择方法的参数,只能自己写一个,代码如下
pca_svd <- function(data, npcs) {
svd_result <- svd(t(data)) # 转置数据矩阵并进行SVD
# 提取前`npcs`个成分
U <- svd_result$u[, 1:npcs]
S <- diag(svd_result$d[1:npcs])
V <- svd_result$v[, 1:npcs]
PCs <- U %*% S # 计算PC得分
return(list("PC" = PCs, "rotation" = V))
}
# 提取Seurat对象中的归一化数据
normalized_data <- GetAssayData(seu, layer = "scale.data")
normalized_data <- normalized_data[VariableFeatures(seu), ]
# 指定要计算的主成分数
npcs <- 50 # 例如,计算前20个PC
# 运行SVD计算PCA
svd_pca_result <- pca_svd(normalized_data, npcs)
# 将结果
embedings <- svd_pca_result$PC
colnames(embedings) <- paste0('PC_', 1:50)
rownames(embedings) <- colnames(seu)
loadings <- svd_pca_result$rotation
colnames(loadings) <- paste0('PC_', 1:50)
rownames(loadings) <- VariableFeatures(seu)
pbmc[["pca"]] <- CreateDimReducObject(
embeddings = embedings,
loadings = loadings,
key = "PC_",
assay = DefaultAssay(pbmc)
)
最后运行之前的SNN构建,UMAP降维和聚类,结果就正常多了。
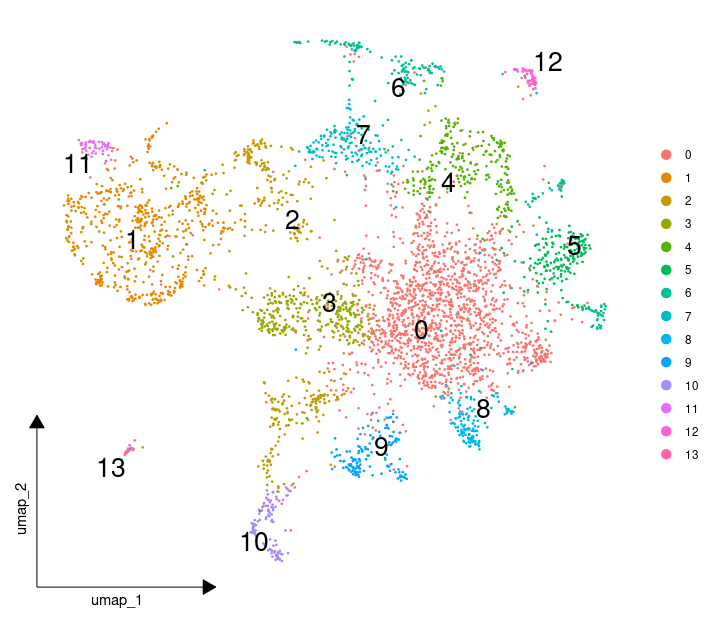
不过,故事还没有结束,因为,我发现相同的代码在Seurat V4是正常的,同时用scanpy的流程也没问题,这个问题就仅仅在Seurat V5中发生。
我感觉,要么安心的留在Seurat V4,要么就大举迁移到Scanpy生态吧。
